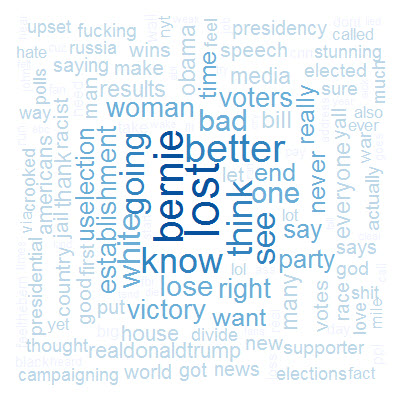
Okay, so I am stoked to report that I can now build them pretty wordclouds ! I am even more pleased with how easy the process is. There’s a whole array of plots you can play around with, including :
Commonality Cloud : Allows you to view words common to both corpora
Comparison Cloud: Allows you view words which are not common to both the corpora
Polarized Plot: A better take on the commonality cloud, allowing you to tell which corpra has a greater concentration of a particular word.
Visualized word Network : shows the network of words associated with a main word.
Let’s jump right into it.
Step 1: Load libraries
require("tm") # the text mining package
require("qdap") # for qdap package's cleaning functions
require("twitteR") # to connect to twitter and extract tweets
require("plotrix") # for the pyramid plot
Step 2: Read in your choice of tweets
After connecting to twitter, I downloaded 5000 tweets each found from a search of the key words “hillary” and “trump”. And this was minutes after the US elections 2016 results were declared . Twitter has never been so lit!
hillary<-searchTwitter("hillary",n=5000,lang = "en")
trump<- searchTwitter("trump",n=5000,lang="en")
Step 3: Write and apply functions to perform data transformation and cleaning
a) Function to extract text from the tweets which get downloaded in the list form.We do this using getText which is an accessor method.
convert_to_text <- function(x){
x$getText()
}
b) Function to process our tweets to remove duplicates and urls.
replacefunc <- function(x){
gsub("https://(.*)", "", x)
}
replace_dup <- function(x){
gsub("^(rt|RT)(.*)", "", x)
}
c) Function to further clean the character vector , for example, to remove brackets, replace abbreviations and symbols with their word equivalents and contractions with their fully expanded versions.
clean_qdap <- function(x){
x<- bracketX(x)
x<- replace_abbreviation(x)
x<- replace_contraction(x)
x<- replace_symbol(x)
x<-tolower(x)
return(x)
}
d) Apply the above functions
hillary_text <- sapply(hillary,convert_to_text)
hillary_text1 <- hillary_text
hill_remove_url<- replacefunc(hillary_text1)
hill_sub <- replace_dup(hill_remove_url)
hill_indx <- which(hill_sub=="")
hill_sub_complete <- hill_sub[-hill_indx]
trump_text <- sapply(trump,convert_to_text)
trump_text1 <- trump_text
trump_remove_url<- replacefunc(trump_text1)
trump_sub <- replace_dup(trump_remove_url)
trump_indx <- which(trump_sub=="")
trump_sub_complete <- trump_sub[-trump_indx]
# encode to UTF-8 : capable of encoding all possible characters defined by unicode
trump_sub_complete <- paste(trump_sub_complete,collapse=" ")
Encoding(trump_sub_complete) <- "UTF-8"
trump_sub_complete <- iconv(trump_sub_complete, "UTF-8", "UTF-8",sub='')
#replace non UTF-8 by empty space
trump_clean <- clean_qdap(trump_sub_complete)
trump_clean1 <- trump_clean
hill_sub_complete <- paste(hill_sub_complete,collapse=" ")
Encoding(hill_sub_complete) <- "UTF-8"
hill_sub_complete <- iconv(hill_sub_complete, "UTF-8", "UTF-8",sub='')
#replace non UTF-8 by empty space
hillary_clean <- clean_qdap(hill_sub_complete)
hillary_clean1 <- hillary_clean
Step 4: Convert the character vectors to VCorpus objects
trump_corpus <- VCorpus(VectorSource(trump_clean1)) hill_corpus <- VCorpus(VectorSource(hillary_clean1))
Step 5: Define and apply function to format the corpus object
clean_corpus <- function(corpus){
corpus <- tm_map(corpus, removePunctuation)
corpus <- tm_map(corpus, stripWhitespace)
corpus <- tm_map(corpus, removeNumbers)
corpus <- tm_map(corpus, content_transformer(tolower))
corpus <- tm_map(corpus, removeWords,
c(stopwords("en"),"supporters","vote","election","like","even","get","will","can"
,"amp","still","just","will","now"))
return(corpus)
}
trump_corpus_clean <- clean_corpus(trump_corpus)
hill_corpus_clean <- clean_corpus(hill_corpus)
- Note : qdap cleaner functions can be used with character vectors, but tm functions need a corpus as input.
Step 6: Convert the corpora into TermDocumentMatrix(TDM) objects
Tdmobjecthillary <- TermDocumentMatrix(hill_corpus_clean1) Tdmobjecttrump <- TermDocumentMatrix(trump_corpus_clean1)
Step 7: Convert the TDM objects into matrices
Tdmobjectmatrixhillary <- as.matrix(Tdmobjecthillary) Tdmobjectmatrixtrump <- as.matrix(Tdmobjecttrump)
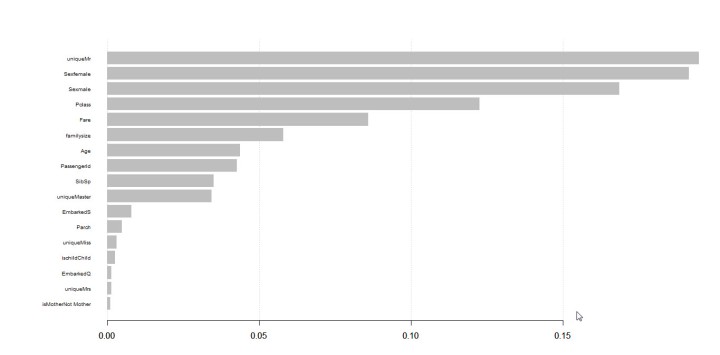
Step 8: Sum rows and create term-frequency dataframe
Freq <- rowSums(Tdmobjectmatrixhillary) Word_freq <- data.frame(term= names(Freq),num=Freq) Freqtrump <- rowSums(Tdmobjectmatrixtrump) Word_freqtrump <- data.frame(term= names(Freqtrump),num=Freqtrump)
Step 9: Prep for fancier wordclouds
# unify the corpora
cc <- c(trump_corpus_clean,hill_corpus_clean)
# convert to TDM
all_tdm <- TermDocumentMatrix(cc)
colnames(all_tdm) <- c("Trump","Hillary")
# convert to matrix
all_m <- as.matrix(all_tdm)
# Create common_words
common_words <- subset(all_tdm_m, all_tdm_m[, 1] > 0 & all_tdm_m[, 2] > 0)
# Create difference
difference <- abs(common_words[, 1] - common_words[, 2])
# Combine common_words and difference
common_words <- cbind(common_words, difference)
# Order the data frame from most differences to least
common_words <- common_words[order(common_words[, 3], decreasing = TRUE), ]
# Create top25_df
top25_df <- data.frame(x = common_words[1:25, 1],
y = common_words[1:25, 2],
labels = rownames(common_words[1:25, ]))
Step 10: It’s word cloud time!
a) The ‘everyday’ cloud
wordcloud(Word_freq$term, Word_freq$num, scale=c(3,0.5),max.words=1000,
random.order=FALSE, rot.per=0.35, use.r.layout=FALSE,
colors=brewer.pal(5, "Blues"))
wordcloud(Word_freqtrump$term, Word_freqtrump$num, scale=c(3,0.5),max.words=1000,
random.order=FALSE, rot.per=0.35, use.r.layout=FALSE,
colors=brewer.pal(5, "Reds"))


b) The Polarized pyramid plot
# Create the pyramid plot
pyramid.plot(top25_df$x, top25_df$y, labels = top25_df$labels,
gap = 70, top.labels = c("Trump", "Words", "Hillary"),
main = "Words in Common", laxlab = NULL,
raxlab = NULL, unit = NULL)

c) The comparison cloud
comparison.cloud(all_m, colors = c("red", "blue"),max.words=100)

d) The commonality cloud
commonality.cloud(all_m, colors = "steelblue1",max.words=100)
We made it! That’s it for this post, folks.
Coming up next: Mining deeper into text.